
MSc student at the University of Glasgow and Glasgow School of Art.
This week the Glasgow School of Art will be holding its 2017 Graduate Degree Show, including work from students completing an MSc in Medical Visualisation and Human Anatomy in a course run jointly with the University of Glasgow. One of these students is Naina Nair, who put her training in medical visualisation to use in constructing the most detailed 3D models currently available of influenza virus particles (virions), using data from the Hutchinson and Bhella groups at the CVR. Here, she writes about how she created models of virions that are as beautiful as they are informative. You might have heard about Naina’s work in the local news, if not, read about it here.
3D visualisations can be used to communicate complex medical and biological processes and in ways that are easier to understand and study. My MSc project was to develop an accurate 3D model of the influenza virion showing the internal structures and also models of the different morphologic forms of the virus especially the filamentous variety (elongated forms; see the CVR’s podcast with Ed Hutchinson on this very topic to learn more).

Data on the viral structure were provided by Dr Hutchinson and his group from the MRC-University of Glasgow Centre for Virus Research. For example, I used mass spectrometry data summarising the abundance of different proteins in the virion (Hutchinson et al. (2014) Nature Communications), combined with structural information to produced by electron microscopy (Vijayakrishnan et al., 2013 PLoS Pathogens) to produce a detailed 3D model. A software workflow of PyMOL or mMaya, MeshLab and 3DsMAx was used to ease the difficulties in importing and converting molecular data which were to be used in developing the models. The process of making these models helped me understand detailed viral data as well as the workings of the different software needed to develop these new visualisations.
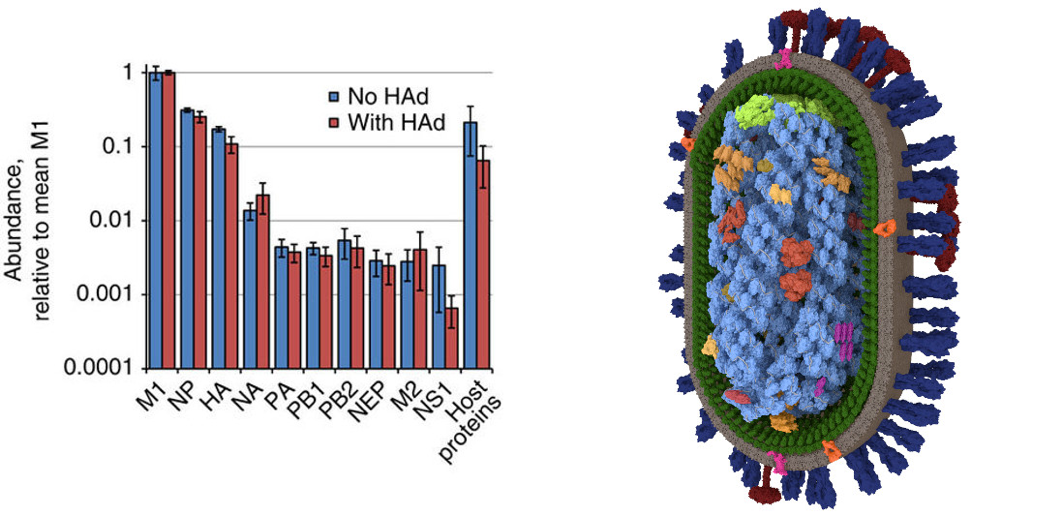
Constant feedback and validation of the processes, by Dr Hutchinson and by the faculty at GSA, were key to developing these models to represent the data as accurately as possible. This project helped develop a new and improved model of the influenza virion in near-atomic detail, as well as what is probably the first 3D model of filamentous influenza virions.
It also helped with formulating some new research related questions which in turn have led to new experiments on the structure of the virus. The development of these models helps bridge the gap between the known and the unknown and creates visualisations which will help the research community with the ongoing study of these infectious viruses.
The Glasgow School of Art Graduate Degree Show is open to the public from the 2 – 7th September, with work from the Simulation and Visualisation course on display in the Reid Building on Renfrew Street. Details at http://www.gsa.ac.uk/life/gsa-events/events/g/graduate-degree-show-2017/
Details about the MSc in Medical Visualisation and Human Anatomy can be found at:
http://www.gla.ac.uk/postgraduate/taught/medicalvisualisation/
http://www.gsa.ac.uk/study/graduate-degrees/medical-visualisation-human-anatomy/

